EVOLUTIONARY GENOMIC ANALYSES OF ADAPTATION IN POPULATIONS
The variation in traits in wild animal and plant populations is one of the most enigmatic features that emerged from natural history studies. Not only do species differ from one another, but also populations of the same species can vary considerably. The insight that natural selection has shaped this observable variation has revolutionized virtually all of biology.
 However, it remains rather uncertain how trait variation is encoded by the genome, and the role of selection on mutations in the genome that matter the most with respect to these traits remains unknown.
However, it remains rather uncertain how trait variation is encoded by the genome, and the role of selection on mutations in the genome that matter the most with respect to these traits remains unknown.
Moreover, we know little if anything regarding the complexity of genetic interactions and how these, in concert, translate into traits. This potentially complex interplay between genes results in difficulties in predicting how selection on genes and their interacting gene-partners could be detected.
Our research is concerned with such detection of selection on genes and their interacting partners. One of our main goals is to map recognizable and ecologically relevant phenotypes onto the genome.
Warfarin resistance in rodent populations
We are studying rat and mouse populations that have evolved a resistance to the anticoagulant rodent poisons warfarin and others. We are searching for genes that confer this adaptive resistance trait and disentangle their potentially complex interactions in genetic networks.
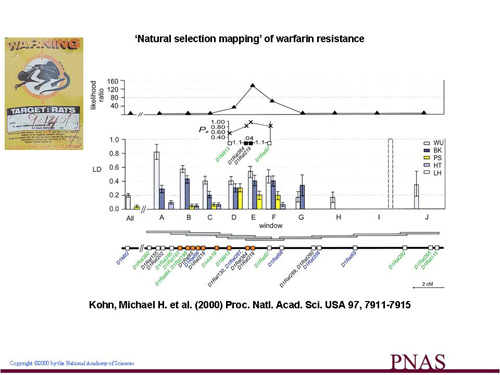
For the main resistance gene, now identified by others as vkorc1, its detailed genomic location can be identified based on the telltale signs of natural selection and genetic hitchhiking. Searches for other genes that contribute to warfarin adaptation could be effectively done in free-living rodent populations in a similar manner, because ~60 years of selection through pest control should have brought any mutations that increase resistance levels (fitness) to a high frequency. We are applying the genomics and bioinformatics toolkit to resistant laboratory strains and wild populations of resistant rats and mice to expand our searches for other warfarin-interacting genes.
What are the functional relationships of the genes, and which pathways are affected? Are all genes involved in the adaptation linked to vitamin K recycling and blood coagulation? Population genetics, network analysis, bioinformatics, in vitro and in vivo studies together should help disentangle the genetic architecture of this adaptation, and to extrapolate from this relatively simple model case study of an adaptation to the evolutionary steps leading towards adaptations in general.
Coat color variation in wild ship rats (Rattus rattus)
Coat color variation is one of few ecologically relevant traits that potentially can be mapped back onto the genotype. This is because decades of genetic research have identified much of the genetic underpinnings of coat coloration. Ship rats occur as at least four coat color morphs, which have been classified as distinct subspecies.
We have begun with the study of the genetic variation underlying this variation in coat coloration. We are examining which genes are associated with the variation, whether the population genetics of any of these can reveal the adaptive nature of the coat color variation, and whether there has been convergent evolution of coat coloration in various locations across the globe.
The genome sequence of this diverse and widespread species has not been sequenced as of yet. We begin with the analysis of genome variation of the R. rattus species complex using next-generation sequencing technologies.