| Reviews
in Undergraduate Research - Issue 1
|
GENETIC
ENGINEERING OF EMBRYONIC STEM CELLS VIA SITE-DIRECTED DNA RECOMBINATION
}
Anna Norman
College of St. Benedict/St. John's University
Dr. Mark MacInnes
Los Alamos National Laboratory, Biosciences Division
|
SUMMARY
This article reviews recent advances in genetic engineering of mammals
utilizing DNA recombination techniques to produce targeted genome modifications.
The general objective of these technologies is to discover novel gene
functions via manipulation of gene expression, regulation, or encoded
protein sequences. The advantage of gene site-directed DNA recombination
is that the engineered variant remains in the normal context of its
chromosomal locus. This feature is especially important for studying
gene function in the context of its regulation in animal development.
The target gene can be subject to both gain of function or loss of function
mutations. In addition to precisely crafted modifications of single
genes, alternatively, site-directed DNA recombination can also produce
chromosomal changes including segment deletion, or inversion, or loss
of heterozygosity between a homologous chromosome pair. Site-directed
recombination is accomplished by certain mechanisms of DNA exchanges
that were first discovered in bacteria and their viruses (bacteriophages).
We will illustrate how these systems permit specific modification of
the mammalian genome. Recombination enzymes of the integrase family
such as the Cre protein (cyclization recombinase) have a well-characterized
site-specific recombination mechanism. Cre recombinase catalyzes DNA
strand exchanges in palindromic DNA target sequences called the locus
of crossover (lox site). The biochemical flexibility of Cre interactions
with lox sites permits a novel approach to mammalian gene targeting.
For example, lox site orientation and-or change of sequences can modify
the specificity of DNA exchange in fascinating ways.
This approach has been most successfully adapted for site-directed recombination
in mouse embryonic stem cells (ES cells). For in vivo analyses,
ES cells can be implanted into embryos to contribute in utero to the
germline tissues. In progeny chimerical mice, the novel genetic trait
may be transmitted via sperm or egg to offspring. ES cells also differentiate
in vitro into numerous cell types allowing direct assessment
of cell lineage phenotypes. Some of the differentiation properties of
mouse ES cells in vitro have been confirmed in both human and
primate ES cells. An understanding of ES cell genetic engineering and
its potential applications is therefore of critical medical and ethical
importance. Despite the successes of these approaches in murine ES cells,
site-directed recombination technology has unresolved questions about
its utility for human genetic and tissue therapy.
DNA RECOMBINATION
IN EMBRYONIC STEM CELLS
In the present review, we will focus on technologies for targeted DNA
recombination in mouse embryonic stem cells (ES cells). These primordial
stem cells are derived from pre-implantation embryos of the blastocyst
stage (Evans and Kaufman, 1981; Martin, 1981). ES cells have the unique
property of pleuripotency, that is the ability of the cells to
differentiate in vivo into all subsequent tissues that arise
from within the embryo (reviewed in Smith, 2001). Upon injection of
ES cells into host blastocyst stage embryos, the ES cells contribute
in utero to embryonic development of all of the somatic and germinal
tissue lineages. A novel line of mice is established when a novel genetic
trait is transmitted in the germline of the founding progeny animals
. To date, there is a literature of several thousand genetically
novel lines of mice created from ES cells.
ES cells also exhibit remarkable proficiency to differentiate in vitro,
providing researchers with many derivative cell types for direct genotype
to phenotype analyses (O'Shea, 1999). Other mammalian cell systems that
are permissive for targeted DNA recombination include chicken DT40 cells
(Dhar et al., 2001; Winding and Berchtold, 2001) and Chinese Hamster
ovary (CHO) cells (Thompson and Schild, 2001). Recently, some of the
characteristics of pleuripotency found mouse ES cells in vitro have
been replicated with both human and monkey ES cell lines (Cibelli et
al., 2002; Reubinoff et al., 2001; Reubinoff et al., 2000). These findings
highlight the crucial medical relevance and ethical implications of
ES cell genetic research regarding the genetic manipulation of human
embryonic stem cell lines.
Complementary Strategies
for Targeted DNA Recombination in ES Cells
Altering target genes within ES cells employs two complementary strategies:
homologous recombination and integrase mechanisms. Homologous recombination
(HR) entails double reciprocal exchanges between DNA molecules dependent
largely on overall sequence identity. HR has long been used to introduce
exogenous DNA sequence into a similar target sequence within mouse
ES cells (Thomas and Cappechi, 1986). In ES cells, HR coupled with
appropriate selection using antibiotics for the desired recombinant
cells provides an efficient method to produce gene-targeted mutations.
Selected cells (clones) derived from single progenitor cells must
be screened for HR by molecular methods (primarily polymerase chain
reaction - PCR) to identify those with the desired genetic modification.
The mechanism of HR within mammals has a complex biochemistry (Thompson
and Schild, 2001). In contrast, prokaryotic DNA recombinases provide
a complementary and less complex avenue towards site-directed genetic
recombination. These mechanisms require much less sequence identity
than HR, decreasing the amount of foreign DNA that is incorporated.
In addition, while the best characterized DNA recombinase enzymes
are of bacterial and yeast origin, they efficiently catalyze recombination
in mammalian cells.
The detailed biochemical understanding of prokaryotic DNA recombinases
and their substrate target sequences has allowed researchers to efficiently
create specific cell type, or developmental control of the timing
of genetic alterations in mice. This feat is accomplished by placing
Cre recombinase under the control of a cell-type-specific gene promoter
with the desired pattern of expression (reviewed in Sauer, 1998).
Another advantage of site-directed DNA recombination is that DNA exchange
is targeted by certain short oligonucleotide DNA sequences (e.g. lox
sites) pre-positioned via HR in the mouse chromosome. In the specific
case of DNA insertion, recombination sites in the plasmid vector identical
to those in the genome provide the substrate for plasmid to chromosome
recombination-mediated insertion of sequence. Variations in lox
site targeting allow repeated exchanges of genetic material at
the same mammalian genome site, or alternatively, into separate sites
in the same genome.
A caveat to this enthusiastic introduction to site directed DNA recombination
is the recent discovery of Cre recombinase-mediated genotoxicity in
the form of chromosome aberrations (Loonstra et al., 2001; Thyagarajan
et al., 2000). Characterization of this untoward consequence of recombinase
activity has led to successful efforts to minimize its confounding
effects on site-directed recombination studies in cells, in vitro
and in tissues, in vivo. We will briefly survey potential applications
of these technologies for human gene therapy in the context of therapeutic
cell type replacement derived from human ES cells. In a related topic
not covered here, others have recently reviewed advances in molecular
methods of assembling gene targeting vectors by HR in E. Coli bacterial
artificial chromosome vectors (BACs) (Copeland et al., 2001).
THE CRE / lox
SITE RECOMBINATION MECHANISM
The integrase family of DNA recombinases shares the biochemical feature
of bimolecular reaction kinetics whereby the enzyme recognizes a specific
DNA recombination sequence. Members of this family include Cre recombinase
from bacteriophage P1, bacteriophage lambda integrase, the yeast Flp
recombinase, and the bacterial XerCD recombinases. These enzymes accomplish
DNA strand exchange in a two step process between DNA substrates. One
pair of strands is exchanged to form a recombination junction intermediate
that does not move, while the second pair of strands is then exchanged
during resolution of the junction. Van Duyne (2001) reviewed the structural
biology of these recombinases with emphasis on the crystal structures
of Cre with its DNA substrate (van Duyne, 2001). Cre recombinase and
to a limited extent Flp recombinase (Seibler et al., 1998) have been
used for enzyme mediated site-directed DNA recombination. We will describe
in some detail how Cre recombinase interaction with its substrate DNA
can be altered in ways that produce diverse genetic outcomes in mammals.
The Cre protein (38 kDa) is encoded by the E. Coli phage P1. P1 is maintained
inside E. Coli cells as a single copy, circular DNA plasmid molecule.
The role of Cre protein is to exchange and separate copies of P1 that
arise after its replication in order to allow partitioning of the two
P1 molecules at each cell division (Hoess and Abremski, 1984; Sternberg
and Hamilton, 1981). The target site of Cre is the loxP sequence
of 34 base-pairs (bp), containing two 13 bp inverted repeats flanking
an 8 bp core sequence (Figure 1). Two Cre molecules bind to each loxP
site, one on each half of the palindrome (van Duyne, 2001). Cre molecules
bound to DNA then form a tetrameric complex bringing two lox
sites into proximity. The Cre-mediated strand cleavage and exchange
between lox sites occurs following the first bases and before
the last base of the 8 bp cores. (The reader is referred to van Duyne
(2001) and references therein for beautiful crystal structure representations
of this complex). The DNA strand asymmetry of the 8 bp core also confers
directionality on the loxP site (Hoess and Abremski, 1984). loxP
orientation determines the type of recombination that will occur between
loxP sites. Cre recombinase catalyzes both inter-molecular DNA
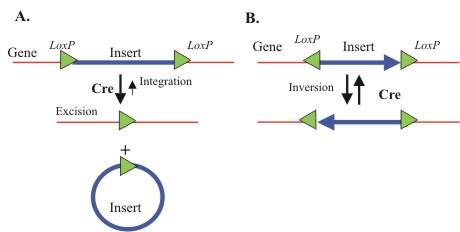
exchanges and intra-molecular excision or inversion (Figure 2). If two
loxP sites in the same molecule are co-aligned, Cre recombination
will preferentially produce excision and circularization of the DNA
between the sites (Figure 2A) (Baubonis and Sauer, 1993; Sauer and Henderson,
1989). Cre also catalyzes the reverse reaction, the integration of DNA
into a single loxP (Figure 2A). However, integration is quite
inefficient since the inserted DNA is immediately flanked by duplicated
loxP sites, which permit re-excision (Araki et al., 1997). When
two loxP sites are inverted in orientation intra-molecular recombination
will produce an orientation switch of the insert (an inversion) with
a 50:50 probability (Figure 2B) (Hoess et al, 1986, Feng et al, 1999).
|
|
Figure
1. DNA sequence of wild type loxP site.
The 13 bp inverted repeats (palindromes) flank an 8 bp asymmetric
core sequence where the recombination exchange takes place. One
Cre recombinase molecule binds to each palindrome sequence (not
shown). Strand cleavage positions are after the first, and before
the last base of the 8-bp core. |
|
|
Figure 2.
Cre mediated loxP recombination reactions at single loxP
sites.
(A) Homologous loxP sites flanking an insert recombine circularize
the insert. B) Recombination between inverted loxP sites
leads to a 50:50 probability of segment inversion. |
The Cre/lox system as outlined above can be used to introduce
certain kinds of gene mutations as well as chromosomal inversions, truncations,
or deletions (Zheng et al., 2000; Feng et al, 1999). Further, Cre-induced
mitotic chromosome recombination between single loxP sites on
each member of a homologue pair has also been used to create genetic
mosaics in mouse ES cells (Liu et al., 2002). This experiment simulates
chromosome loss of heterozygosity (LOH) that is seen in many types of
tumors. Cre-induced mitotic recombination in a tissue lineage in
vivo would permit studies of LOH effects on development or in adult
mouse tissues.
The recombination properties of Cre at a single loxP site select
against the insertion of precise DNA segments into the target chromosome.
Upon targeted integration of DNA, the loxP site is duplicated,
leading to the highly favored intra-molecular excision (Fig. 2A). When
an integration event does occur, not only is the DNA of interest integrated
into the genome, DNA from the targeting plasmid vector is integrated
as well. Alternative strategies have been devised using the Cre/lox
system in order to create higher frequency and stability of insertion
events, in vitro, and ultimately to eliminate plasmid DNA introduction
into the genome.
CRE RECOMBINATION
WITH MUTANT lox SITES: STRATEGIES FOR DNA TO CHROMOSOME INSERTION
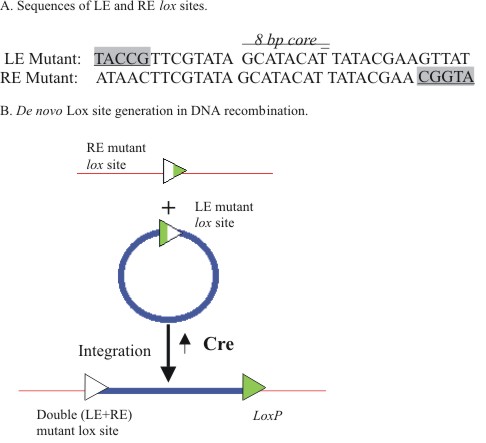
Albert et al. (1995) found that mutations in loxP permit integration
of DNA at a plant target site in the plant genome while avoiding its
immediate re-excision. This strategy was also successful for integrating
foreign gene DNA into a mouse chromosome (Araki et al., 1997). A single
mutant lox site in which nucleotides were altered in the right
hand palindrome was pre-positioned by HR in the chromosome target (Figure
3). In the targeting vector, a distinct lox mutation was incorporated
into the left hand palindromic element. The two mutant lox sites
were in co-alignment and still enabled Cre to catalyze inter-molecular
recombination in ES cells. However, the integration resulted in the
creation of two de novo lox sites, one containing both left-end
and right-end mutations, while normal lox P was generated at
the other site. Cre poorly recognizes the LE+RE lox site, which
inhibited re-excision between it and the loxP. This approach
facilitated efficient insertion of a targeting cassette.
|
|
Figure 3.
The LE/RE mutant lox site strategy for segment integration.
Cre-mediated recombination between the mutant right end (RE) and
left end (LE) lox sites produces a trapped product double
mutant (LE+RE) lox site and a WT loxP site that are
less susceptible to intra-molecular excision. |
The idea that different lox sites may not recombine efficiently
but that identical lox site recombination remains proficient
led to an in-depth study of these interactions. Lee and Saito (1998)
identified many mutant lox sites that recombine efficiently with
an identical partner complex but not with loxP (Figure 4A). For
example, lox 2272 and lox 2372 sequences contain two nucleotide
changes in the core 8-bp sequence (Lee and Saito, 1998). The lox
FAS site occurs naturally in Saccharomyces cerevisiae (Sauer,
1996). Lox FAS has a completely different consensus core sequence
from loxP while remaining an efficient substrate for Cre. This
fact illustrates the plasticity of lox sites. The lox 511
site contains a single nucleotide mutation in the core sequence (Hoess
et al., 1986). The recombination efficiency between homologous and heterologous
pairs of lox sites has been studied in E. Coli (Siegel
et al., 2001) (Table 1). Their results show that these heterologous
pairs of lox sites undergo recombination at a much lower frequency
than homologous pairs.
| Table
1. Recombination frequencies in E. coli among three mutant
lox sites and loxP. Reprinted with permission of the authors. Note,
Siegel et al (2001) reported recombination results for a sequence
originally thought to be lox2272. However, upon inspection
the published sequence it was not lox2272 but rather lox2372
(from Lee and Saito, 1998). (A. Bradbury, personal communication) |
|
.
|
WT
|
2272
|
FAS
|
511
|
|
WT
|
99.6
|
.
|
.
|
.
|
|
2272
|
0.5
|
99.7
|
.
|
.
|
|
FAS
|
0.2
|
1.7
|
99.4
|
.
|
|
511
|
10.3
|
1.6
|
0.0
|
99.8
|
Siegel et al. (2001) used
a green used fluorescent protein (GFP) gene flanked by heterologous
lox sites in the lac Z reporter gene (responsible for
production of b-galactosidase) of a plasmid
DNA. Before recombination, the bacterial colonies expressed GFP and
emitted green fluorescence. Correct recombination events resulted in
excision of the GFP gene and permitted the lac Z gene to be translated,
producing a loss of GFP fluorescence and concomitant expression of functional
lac Z gene. Such E. Coli recombinant colonies were non-fluorescent
and blue dye colored on X-gal medium. The assay quantified the accumulated
recombination events over many generations of colony growth (~18h) and
therefore it was very sensitive to low levels of correct recombination.
The assay also distinguished aborted or aberrant recombination repair
products from true GFP excision via absence of correct lacZ gene
activation. These findings indicated that recombination between homologous
pairs of lox sites (whether similar or dissimilar to loxP)
can occur efficiently in vivo while recombination between heterologous
pairs occurs much less efficiently. Lee and Saito (1998) also noted
the occurrence in some combinations of arrested intermediate recombination
structures in their in vitro plasmid assay system. In these situations,
recombination proceeded to exchange one DNA strand but, due to the heterozygosity
of the lox sites, the intermediate wasn't able to resolve into
the final recombination product. The potential persistence of arrested
intermediates between heterologous lox sites may have implications
for the use of the Cre/lox system in mammals (see below).
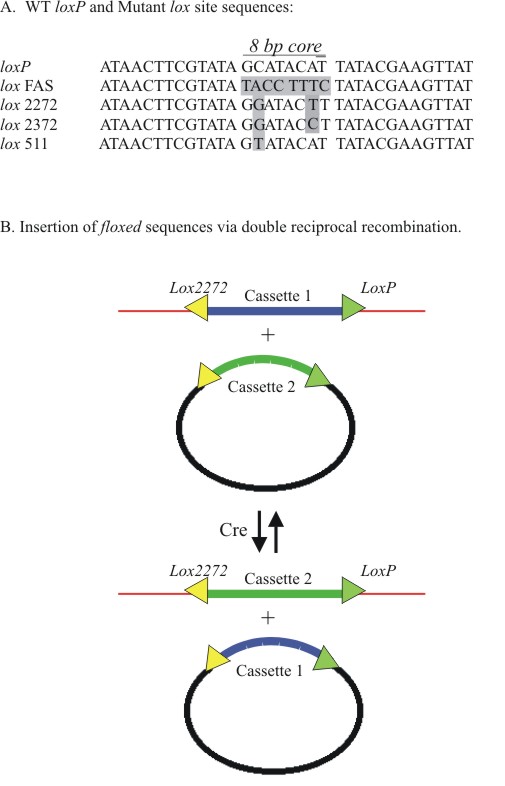 |
Figure
4. Recombinase Mediated Cassette Exchange (RMCE).
A) Sequence differences in mutant lox sites. B) Heterologous
lox sites (in this example loxP and lox 2272)
sites flanking an insert can be used to swap pre-placed genome cassette
(Cassette 1) for a targeting insert (Cassette 2) that is flanked
by the same lox sites. Recombination events that result in
successful cassette exchange are assessed by molecular analysis
or by antibiotic growth selection. |
Given the low level of Cre-mediated recombination between several heterologous
pairs of lox sites, new gene targeting techniques were developed that
exploit this selectivity. The DNA to be inserted into the genome is
constructed so that it is flanked by heterologous lox sites. The genomic
target contains the matching lox sites by HR pre-placement. In the presence
of Cre, during a double-reciprocal recombination event, there occurs
a 50:50 probability of replacement of the lox-flanked chromosomal DNA
by the targeting allele (Figure 4B). This exchange is referred to as
recombinase-mediated cassette exchange, or RMCE. The RMCE system permits
efficient insertion of lox-flanked DNA into the mammalian genome (Feng
et al., 1999; Kolb, 2001; Trinh and Morrison, 2000). It is used to swap
wild-type functional gene segments with knockout or otherwise mutated
gene segments without incorporation of extraneous DNA.
Kolb used HR and site-specific RMCE to successfully insert a reporter
gene into the mouse b-casein locus (Kolb,
2001). Kolb created a targeting construct consisting of lox 2272
and loxP sites flanking a selection marker that was integrated
into the ES cells genome via HR. Recombination with a targeting construct
containing a luciferase reporter gene flanked by lox2272 and
loxP sites resulted in the efficient switching of the lox-flanked
cassettes (Figure 4B). Typically, the HR step involves using selection
markers such as geneticin (G418) or hygromycin resistance for positive
selection placement of the lox-flanked gene segment. In
RMCE, the selection marker is removed to avoid dysregulation of the
modified allele. The loss of the selection marker by site directed recombination
is tested by replica plating of cell clones in the appropriate selection
medium.
CRE RECOMBINASE
EXPRESSION: REGULATION IN MAMMALIAN DEVELOPMENT
The Cre/lox system for genetic recombination also permits lox-flanked
target gene alteration via stage- or tissue-specific control dependent
upon the regulation of Cre gene expression in vivo. The majority
of mammalian genes are thought to have developmentally regulated expression
or they may express only in specific tissues. Because lox sites
are quite short, their presence in the genome does not generally impair
expression of their 'host' gene (Silver and Livingston, 2001; Trinh
and Morrison, 2000). Often, expression of a mutation in the whole animal
rather than at a specific time or tissue location would prove lethal,
thereby preventing the study of phenotypes of the gene (Schipani, et
al., 2001). The targeted gene is flanked by loxP sites and then
integrated into the genome via HR. The altered ES cells are then developed
into mice with the lox-flanked gene intact. The Cre gene is engineered
to express under the control of a cell-type-specific promoter, whichever
suits the purpose of the study (Metzger and Chambon, 2001; Metzger and
Feil, 1999; Schipani et al., 2001). A line of Cre tissue-specific expression
transgenic mice is created separately and evaluated for cell-type specific
Cre-expression. When the two transgenic mouse lines are mated, the progeny
of doubly transgenic genotype will enable activation of Cre expression
in the appropriate cell type or time of development. The mutation (usually
an excision recombination) arises by Cre expression in most of the affected
cells. Typically at low levels of Cre expression, some cells are mutated
while others are not, creating genetic mosaicism in the tissue. Mosaicism
may be useful for phenotype interpretation by providing modified and
unmodified cells side by side.
In addition, it is important to assess whether Cre may cause untoward
effects such as cell death arising from Cre expression in the transgenic
parental and non-targeted littermate mice. Many transgenic mouse lines
thought to have tissue-specific Cre-transgene expression appear normal
outwardly and by histology (Lewandoski, 2001; Nagy and Mar, 2001). We
shall see next why careful examination is warranted of tissue specific
Cre-transgene expression.
CRE RECOMBINASE
GENOTOXICITY
While site-directed recombination is a useful tool for genetic manipulation,
Cre recombinase is also inherently toxic to many mammalian cells lines.
This toxicity is the result of the recombinase activity of Cre (Loonstra
et al., 2001; Silver and Livingston, 2001). These researchers have reported
total cessation of cell replication, cell death, and an abundance of
chromosomal aberrations and aneuploidy following high level Cre
recombinase expression. These events could be the result of illegitimate
DNA recombination or strand breaks induced in the mammalian genome.
Another observation consistent with this notion is that cells cultured
in the presence of high levels of Cre showed an increase in the number
of cells in the G2/M phase of the cell cycle (the period just before
mitosis begins and mitosis itself). This result indicates that the DNA
damage is severe enough to trigger marked G2/M cell cycle checkpoint
arrest (for a RUR review of cell cycle check points, see the article
by Renthal in this issue).
A corroborating discovery was that of the existence of pseudo-lox sites
in the mammalian genome (Thyagarajan et al., 2000). Illegitimate mammalian
genomic lox sites elicited Cre-mediated recombination. Indeed, Cre also
induces recombination at secondary recombination sites that occur naturally
in E. Coli and in yeast (called loxB sites) (Sauer, 1996; Sternberg
and Hamilton, 1981). As Cre catalyzes apparent interaction between pseudo-lox
sites in mammalian cells, these events could therefore result in deletions
or other chromosome alterations. Consequently, Cre induced breaks at
endogenous chromosomal sites may possibly complicate the interpretation
of Cre/lox experiments.
The studies that elucidated this problem also offered possible technical
solutions. Loonstra et al. used a hormone-regulated Cre gene that was
expressed at negligible levels without induction (Loonstra et al., 2001).
When cells were subjected to supra-basal but not saturating levels of
the hormone, Cre expression was elevated sufficiently to catalyze excision
of a lox-flanked reporter gene without inhibiting cell growth
and without production of visible chromosome aberrations. In their work
cited in Loonstra et al (2001) a moderate level of the hormone elicited
complete excision of another lox flanked genomic target without
apparent genotoxicity. Others have employed variations on 'hit and run'
strategies utilizing a negative feedback loop to circumvent overt Cre
genotoxicity (Pfeifer et al., 2001; Silver and Livingston, 2001). Cre
expression vectors were engineered to produce low levels of Cre coupled
with a genetic negative feedback loop to limit the amount of Cre in
the cells. Silver and Livingston used a retroviral vector containing
a Cre expression gene with a single lox 511 site in its LTR.
This retroviral vector was engineered so that following its reverse
transcription and genome integration, the Cre-expression vector contained
duplicated LTRs with co-aligned lox 511 sites. When Cre was expressed
at a level high enough to cause recombination between the lox
511 sites, the entire Cre gene was auto-excised removing further synthesis
of Cre after a few cell generations. This strategy resulted in cells
capable of targeted excision of lox-flanked sequences in an unlinked
target gene. As Cre expression was limited, there were no observed genotoxic
effects.
It is also likely that the amount of Cre expressed in ES cells in culture
can be controlled simply during the gene transfer process. Linear DNA
introduced into ES cells by the technique of electroporation is efficiently
integrated into the genome, either randomly or by HR, in ES cells. In
contrast, circular plasmid DNA has approximately 8 fold lower probability
of chromosome integration (Taniguchi et al., 1998). This difference
can be exploited to control magnitude and duration of Cre-recombinase
expression simply via the transient presence of Cre expression plasmid.
In addition, a fluorescence reporter plasmid called 'Cre-Stoplight'
has been developed recently to bioassay Cre recombinase activity in
live cells by epifluorescence microscopy or flow cytometry (Yang and
Hughes, 2001). The plasmid incorporates dual reporter gene cassettes
containing a lox-flanked Discosoma coral fluorescent protein,
DsRed, and a transcriptionally inactive green fluorescent protein (EGFP).
When sufficient Cre is expressed in cells containing Cre-Stoplight the
DsRed gene is excised and rendered inactive by virtue of its flanking
lox sites. Then the upstream promoter is brought into apposition
to the EGFP gene. Therefore, mouse ES cells taking up DNA after 72 hours
show considerable fractions of cells (> 20 %) with both red and green
epifluorescence caused by the switch of DsRed to GFP production (K.
Nowak and M. MacInnes, unpublished observations). We are now investigating
the utility of monitoring levels of Cre recombinase by transient expression
of Cre Stoplight to obtain efficient GFP activation, and presumably
recombination at specific genomic target sites. As indicated above,
for in vivo experiments similar engineering of tissue-specific
autoexcision, or autoregulation, of Cre transgene may help avoid the
possibility of confounding non-specific genotoxicity in the developmental
stage or tissue of interest.
SITE-DIRECTED RECOMBINATION
IN MAMMALIAN FUNCTIONAL GENOMICS AND HUMAN GENE THERAPY
The Cre/lox recombination system and HR have given researchers powerful
tools for investigating novel mammalian genes. Conversely, they can
also be used to create controlled gross deletions, inversions, and chromosome
mitotic recombination in order to characterize certain genetic disease
processes. The rich applications of Cre/lox hold promise for elucidating
thousands of novel gene functions, an essential integrative genetics
component of the functional genomics / systems biology era. The recent
completion of the draft sequences of both mouse and human genomes will
greatly facilitate building HR and Cre/lox recombination vectors in
both mouse and human cells. The production of gene targeting vectors
for HR and Cre/lox strategies must be automated into a high-throughput
enterprise in order to realize the full potential of these approaches
(Copeland et al., 2001). Characterization of gene function in ES cells
and their derivatives in vitro would facilitate preliminary genetic
analyses without necessitating very costly and ethically questionable
production of tens of thousands of new mutant mouse lines.
It is of great interest whether HR gene targeting is possible in human
ES cells given that few or no diploid human cell lines have yet proved
useful for HR and site-directed recombination. Recent provocative research
produced successful isolation of stem-like cells from human and monkey
embryos (Reubinoff et al., 2001; Reubinoff et al., 2000; Thomson and
Marshall, 1998) and from parthenogenetically activated Macac eggs (Cibelli
et al., 2002). Similarly, adult stem cells have been isolated from mammalian
bone marrow, liver, pancreas and brain (for a review see Clarke and
Frisen, 2001). These milestones raise controversial ethical possibilities
that human cell therapy (and huES cell genetic engineering) may become
a reality for numerous diseases with a genetic component. In theory,
human stem cells could be 'corrected' through retroviral vector incorporation
or via HR, and this approach complemented by Cre/lox genetics. Two major
technical concerns in cell replacement therapy are, first, the possibility
of implanted cell/tissue rejection. Ideally, this difficulty is circumvented
by use of the patient's own (autologous) stem cells. The second technical
problem concerns a significant possibility of a carcinoma arising from
implanted stem-like cells. As illustrated in this brief review, we have
shown how activation or reversal of targeted genetic modifications can
be engineered using Cre/lox. This approach may offer opportunities to
provide additional safeguards against neoplasia in therapeutic strategies
involving cell replacement with human stem cells.
ABOUT THE AUTHOR
Anna Norman is currently a junior at the College of St. Benedict in
St. Joseph, Minnesota where she is majoring in Biology. After graduation,
she hopes to attend either medical school or to enroll in a joint MD
/ PhD program. During the summer of 2001, she was accepted to a National
Science Foundation sponsored Research Experience for Undergraduate students
(REU) Program hosted by the Los Alamos National Laboratory. Under the
mentorship of Dr. Mark A. MacInnes, a geneticist in Biosciences Division,
she tested recombination efficiency of a homologous and heterologous
pair of lox sites flanking a marker gene introduced into mouse
ES cells. Using recombinant DNA techniques, Ms. Norman assembled the
plasmid vectors containing pairs of lox sites flanking a drug
resistance marker gene. These vectors were then linearized and introduced
into mouse ES via electroporation. Recombination efficiency was analyzed
based on the number of colonies that grew, and cell growth rate, in
culture medium containing antibiotics. Using this method she found resistance
to intra-molecular recombination between lox 2372 and lox
FAS but proficient as recombination as expected between loxP
sites. During the experiment, quantifying colony yield and cell regrowth
confirmed that certain toxic effects of Cre recombinase occurred in
mouse ES cells leading in part to the discussion of Cre genotoxicity
in this review. Improvement of methods outlined in the review is a major
emphasis of the MacInnes laboratory at this time.
FURTHER READING
Metzger, D. and Feil, R. (1999). Engineering the mouse genome by site-specific
recombination. Curr. Opin. Biotechnol. 10, 470-476.
Sauer, B. (1998). Inducible Gene Targeting in Mice Using the Cre/lox
System. Methods 14, 381-392.
REFERENCES
Albert, H., Dale, E. C.,
Lee, E., and Ow, D. W. (1995). Site-specific integration of DNA into
wild-type and mutant lox sites placed in the plant genome. Plant
J 7, 649-659.
Araki, K., Araki, M., and
Yamamura, K. (1997). Targeted integration of DNA using mutant lox sites
in embryonic stem cells. Nucleic Acids Res 25, 868-872.
Araki, K., Imaizumi, T.,
Sekimoto, T., Yoshinobu, K., Yoshimuta, J., Akizuki, M., Miura, K.,
Araki, M., and Yamamura, K. (1999). Exchangeable gene trap using the
Cre/mutated lox system. Cell Mol Biol (Noisy-le-grand) 45,
737-750.
Baubonis, W., and Sauer,
B. (1993). Genomic Targeting with Purified Cre Recombinase. Nucleic
Acids Res 21, 2025-2029.
Cibelli, J. B., Grant, K.
A., Chapman, K. B., Cunniff, K., Worst, T., Green, H. L., Walker, S.
J., Gutin, P. H., Vilner, L., Tabar, V., et al. (2002). Parthenogenetic
stem cells in nonhuman primates. Science 295, 819.
Clarke, D., and Frisen, J.
(2001). Differentiation potential of adult stem cells. Curr Opin
Genet Dev 11, 575-580.
Copeland, N. G., Jenkins,
N. A., and Court, D. L. (2001). Recombineering: a powerful new tool
for mouse functional genomics. Nat Rev Genet 2, 769-779.
Dhar, P. K., Sonoda, E., Fujimori, A., Yamashita, Y. M., and Takeda,
S. (2001). DNA repair studies: experimental evidence in support of chicken
DT40 cell line as a unique model. J Environ Pathol Toxicol Oncol
20, 273-283.
Evans, M. J., and Kaufman,
M. H. (1981). Establishment in culture of pluripotential cells from
mouse embryos. Nature 292, 154-156.
Feng, Y.-Q., Seibler, J.,
Alami, R., Eisen, A., Westerman, K. A., Leboulch, P., Fiering, S., and
Bouhassira, E. E. (1999). Site-specific Chromosomal Integration in Mammalian
Cells: Highly Efficient CRE Recombinase-mediated Cassette Exchange.
J Mol Biol 292, 779-785.
Hoess, R. H., and Abremski,
K. (1984). Interaction of the bacteriophage P1 recombinase Cre with
the recombining site P1. Proc Natl Acad Sci USA 81, 1026-1029.
Hoess, R. H., Wierzbicki,
A., and Abremski, K. (1986). The role of the loxP spacer region
in P1 site-specific recombination. Nucleic Acids Res 14,
2287-2330.
Kolb, A. F. (2001). Selection-marker-free
modification of the murine beta-casein gene using a lox2272 [correction
of lox2722] site. Anal Biochem 290, 260-271.
Lee, G., and Saito, I. (1998).
Role of nucleotide sequences of loxP spacer region in Cre-mediated
recombination. Gene 216, 55-65.
Lewandoski, M. (2001). Conditional
control of gene expression in the mouse. Nat Rev Genet 2,
743-755.
Liu, P. T., Jenkins, N. A.,
and Copeland, N. G. (2002). Efficient Cre-loxP-induced mitotic
recombination in mouse embryonic stem cells. Nature Genetics
30, 66-72.
Loonstra, A., Vooijs, M.,
Beverloo, H. B., Allak, B. A., van Drunen, E., Kanaar, R., Berns, A.,
and Jonkers, J. (2001). Growth inhibition and DNA damage induced by
Cre recombinase in mammalian cells. Proc Natl Acad Sci USA 98,
9209-9214.
Martin, G. R. (1981). Isolation
of a pluripotent cell line from early mouse embryos cultured in medium
conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA
78, 7634-7638.
Metzger, D., and Chambon,
P. (2001). Site- and Time-Specific Gene Targeting in the Mouse. Methods
24, 71-80.
Metzger, D., and Feil, R.
(1999). Engineering the mouse genome by site-specific recombination.
Curr Opin Biotechnol 10, 470-476.
Nagy, A., and Mar, L. (2001).
Creation and use of a Cre recombinase transgenic database. Methods
Mol Biol 158, 95-106.
O'Shea, K. S. (1999). Embryonic
stem cell models of development [see comments]. Anat Rec 257,
32-41.
Pfeifer, A., Brandon, E.
P., Kootstra, N., Gage, F. H., and Verma, I. M. (2001). Delivery of
the Cre recombinase by a self-deleting lentiviral vector: Efficient
gene targeting in vivo. Proc Natl Acad Sci USA 98, 11450-11455.
Reubinoff, B. E., Itsykson,
P., Turetsky, T., Pera, M. F., Reinhartz, E., Itzik, A., and Ben-Hur,
T. (2001). Neural progenitors from human embryonic stem cells. Nat
Biotechnol 19, 1134-1140.
Reubinoff, B. E., Pera, M.
F., Fong, C. Y., Trounson, A., and Bongso, A. (2000). Embryonic stem
cell lines from human blastocysts: somatic differentiation in vitro.
Nat Biotechnol 18, 399-404.
Sauer, B. (1996). Multiplex
Cre/lox recombination permits selective site-specific DNA targeting
to both a natural and an engineered site in the yeast genome. Nucleic
Acids Res 24, 4608-4613.
Sauer, B. (1998). Inducible
Gene Targeting in Mice Using the Cre/lox System. Methods 14,
381-392.
Sauer, B., and Henderson,
N. (1989). Cre-stimulated recombination at loxP-containing DNA
sequences placed into the mammalian genome. Nucleic Acids Res.
Seibler, J., Schubeler, D.,
Fiering, S., Groudine, M., and Bode, J. (1998). DNA cassette exchange
in ES cells mediated by Flp recombinase: an efficient strategy for repeated
modification of tagged loci by marker-free constructs. Biochemistry
37, 6229-6234.
Siegel, R. W., Jain, R.,
and Bradbury, A. (2001). Using an in vivo phagemid system to
identify non-compatible loxP sequences. FEBS Let 499,
147-153.
Silver, D. P., and Livingston,
D. M. (2001). Self-Excising Retroviral Vectors Encoding the Cre Recombinase
Overcome Cre-Mediated Cellular Toxicity. Mol Cell 8, 233-243.
Smith, A. G. (2001). Embryo-derived
stem cells: of mice and men. Annu Rev Cell Dev Biol 17,
435-462.
Sternberg, N., and Hamilton,
D. (1981). Bacteriophage P1 Site-Specific Recombination I. Recombination
Between loxP Sites. J Mol Biol 150, 467-507.
Taniguchi, M., Sanbo, M.,
Watanabe, S., Naruse, I., Mishina, M., and Yagi, T. (1998). Efficient
production of Cre-mediated site-directed recombinants through the utilization
of the puromycin resistance gene, pac: a transient gene-integration
marker for ES cells. Nucleic Acids Res 26, 679-680.
Thomas, K. R., and Capecchi,
M. R. (1986). Introduction of homologous DNA sequences into mammalian
cells induces mutations in the cognate gene. Nature 324,
34-8.
Thomson, J. A., and Marshall,
V. S. (1998). Primate embryonic stem cells. Curr Top Dev Biol 38,
133-65.
Thompson, L. H., and Schild,
D. (2001). Homologous recombinational repair of DNA ensures mammalian
chromosome stability. Mutat Res 477, 131-153.
Thyagarajan, B., Guimaraes,
M. J., Groth, A. C., and Calos, M. P. (2000). Mammalian genomes contain
active recombinase recognition sites. Gene 244, 47-54.
Trinh, K. R., and Morrison,
S. L. (2000). Site-specific and directional gene replacement mediated
by Cre recombinase. J Immunol Methods 244, 185-193.
van Duyne, G. D. (2001).
A Structural View of Cre-loxP Site-Specific Recombination. Annu
Rev Biophys Biomol Struct 30, 87-104.
Winding, P., and Berchtold,
M. W. (2001). The chicken B cell line DT40: a novel tool for gene disruption
experiments. J Immunol Methods 249, 1-16.
Yang, Y. S., and Hughes,
T. E. (2001). Cre stoplight: a red/green fluorescent reporter of Cre
recombinase expression in living cells. Biotechniques 31,
1036, 1038, 1040-1031.
Zheng, B., Sage, M., Sheppeard,
E. A., Jurecic, V., and Bradley, A. (2000). Engineering Mouse Chromosomes
with Cre-loxP: Range, Efficiency, and Somatic Applications. Mol
Cell Biol 20, 648-655.
|