Metabolic engineering is the
application of engineering design and analysis to the metabolic pathways of
cells in order to achieve a particular goal such as to increase process
productivity or to extend metabolic capability of cells by the addition of
extrinsic activities for chemical production or degradation. In our lab we have
exploited several different molecular biology techniques to improve cellular
properties and to study the relation between cellular function and genotype.
Characterization of genetically engineered strains is then performed with the
use of bioreactors
and analytical techniques such as HPLC are used to measure the metabolites produced. One of the
most novel metabolic engineering tools developed in our lab, cofactor
manipulations, has been used to investigate the importance of maintaining a
proper balance of the biosynthetic transformations involving
oxidation-reduction reactions in the cell. There are infinite applications of
metabolic engineering in the medical and the industrial fields. Currently our
group is investigating the following topics:
- Cofactor manipulations in Escheria coli
- Manipulation of NADH availability
- Acetyl-CoA manipulation
- Biochemical patyway manipulation
- Plant metabolic engineering
- Quantative biotechnology
Cofactor
manipulations in Escherichia
coli
Most current metabolic
engineering studies have focused on manipulations of enzyme levels and the
effect of amplification, addition or deletion of a particular pathway. A newer
focal area is the manipulation of cofactors, which play a major role in cell
physiology and in the production of a number of fermentation products.
Additionally they are essential for enzymes and serve as donor and/or acceptors
of electrons so enzymes can catalyze biological redox reactions. Part of the
research in our lab is to investigate the effect of different cofactor
manipulations using cofactor-dependent products as a model system.
Manipulation
of NADH availability
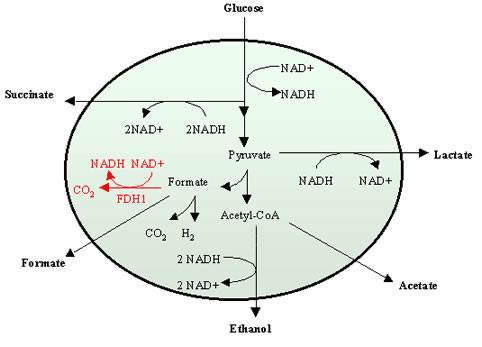
The NADH/NAD+ cofactor pair plays a major role in microbial catabolism, in which a carbon source such as glucose is oxidized utilizing NAD+ as a cofactor and produces reducing equivalents in the form of NADH. The reducing power is subsequently used for the synthesis of fermentative products. In the case of succinate production in Escherichia coli under anaerobic conditions, two moles of NADH are required to form one mole of succinate. However, under anaerobic conditions, one mole of glucose can only provide two moles of NADH. Thus, this cofactor is the limiting agent to achieve the maximum theoretical yield of succinate from glucose.
We are currently working with a new in vivo NADH recycling system to increase the intracellular NADH availability and applying this system for the production of succinate. This system illustrates that cofactor manipulation can be used as an additional tool to further increase succinate productivity.
Schematic representation of the central anaerobic metabolic pathways of E. coli showing involvement of the NADH/NAD+ cofactor pair and the newly introduced NAD+-dependent pathway (shown in red).
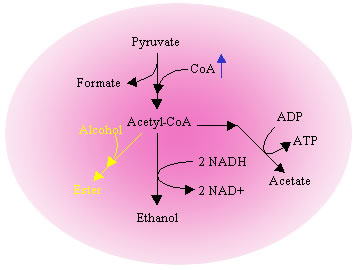
Another important cofactor combination is CoA and its derivative acetyl-CoA. Acetyl-CoA is an essential intermediate in many energy yielding metabolic pathways and a substrate in enzymatic production of industrially useful compounds such as esters. The purpose of this project is to investigate the metabolic flux redistribution in E. coli upon precise perturbations of CoA levels. The production of chemicals that require acetyl-CoA such as isoamyl acetate has been used as a model system to demonstrate the beneficial effects of altering CoA metabolism.
Ester production pathway with increased CoA levels
Biochemical
pathway manipulation
Succinic acid is a chemical with broad industrial value. It acts as a potential precursor to many commercial chemicals. Traditionally, succinic acid is produced through chemical processes. In the last few years, efforts have shifted toward microbial fermentation for production. We have been trying to genetically engineer E. coli strains to produce succinic acid as a major fermentation product by manipulating different important enzymes involved in the aerobic and anaerobic production of succinate.
In a joint project with Dr. Jacqueline Vanni Shanks of Iowa State University and Dr. Sue Gibson, our lab explores the area of metabolic engineering in plants. We have developed transgenic "hairy root" cultures of the periwinkle Catharanthus roseus. Hairy root cultures are biochemically stable and have provided an excellent system for metabolic studies for the production of valuable natural plant products.
In our laboratories, we are interested in the metabolic engineering of the indole pathway and its subsequent effect on indole alkaloid biosynthesis in Catharanthus roseus hairy roots. C. roseus produces a wide variety of indole alkaloids including the anti-cancer drugs vincristine and vinblastine. Hairy roots have a number of advantages over cell cultures including increased genetic stability and substantially higher alkaloid content.
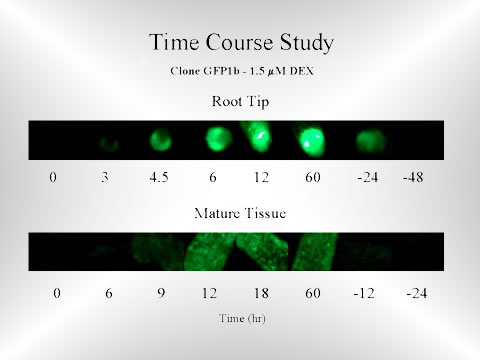
Time course of dexamethasone-inducible GFP expression in transgenic C. roseus hairy roots. Mature regions and tips are represented with negative hours indicating time after the removal of dexamethasone.
Recent rapid accumulation of
enormous amount of genomic and pathway information provides a unique
opportunity for the optimal design of genetic circuits and/or metabolic
pathways to achieve a specific cellular behavior. Our laboratory is interested
in the development of new design approaches and tools for the efficient
utilization of these massive genomic and pathway knowledge.

Rice
University
Department of Bioengineering
6100 Main
Houston, TX 77005
Office: BRC 815
Lab: BRC 820